Abstract
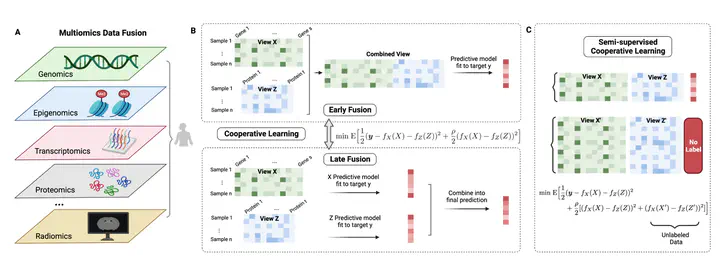
Multiomics data fusion integrates diverse data modalities, ranging from transcriptomics to proteomics, to gain a comprehensive understanding of biological systems and enhance predictions on outcomes of interest related to disease phenotypes and treatment responses. Cooperative learning, a recently proposed method, unifies the commonly-used fusion approaches, including early and late fusion, and offers a systematic framework for leveraging the shared underlying relationships across omics to strengthen signals. However, the challenge of acquiring large-scale labeled data remains, and there are cases where multiomics data are available but in the absence of annotated labels. To harness the potential of unlabeled multiomcis data, we introduce semi-supervised cooperative learning. By utilizing an “agreement penalty”, our method incorporates the additional unlabeled data in the learning process and achieves consistently superior predictive performance on simulated data and a real multiomics study of aging. It offers an effective solution to multiomics data fusion in settings with both labeled and unlabeled data and maximizes the utility of available data resources, with the potential of significantly improving predictive models for diagnostics and therapeutics in an increasingly multiomics world.