LipidCCS prediction of collision cross-section values for lipids with high precision to support ion mobility–mass spectrometry-based lipidomics
Abstract
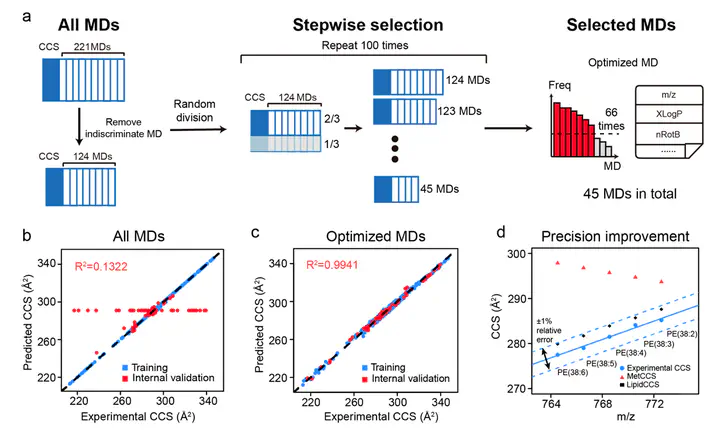
The use of collision cross-section (CCS) values derived from ion mobility–mass spectrometry (IM–MS) has been proven to facilitate lipid identifications. Its utility is restricted by the limited availability of CCS values. Recently, the machine-learning algorithm-based prediction (e.g., MetCCS) is reported to generate CCS values in a large-scale. However, the prediction precision is not sufficient to differentiate lipids due to their high structural similarities and subtle differences on CCS values. To address this challenge, we developed a new approach, namely, LipidCCS, to precisely predict lipid CCS values. In LipidCCS, a set of molecular descriptors were optimized using bioinformatic approaches to comprehensively describe the subtle structure differences for lipids. The use of optimized molecular descriptors together with a large set of standard CCS values for lipids (458 in total) to build the prediction model significantly …