KEGG pathway enrichment
Xiaotao Shen PhD (https://www.shenxt.info/)
true
Created on 2020-03-28 and updated on 2021-03-13
Source:vignettes/kegg_pathway_enrichment.Rmd
kegg_pathway_enrichment.Rmd
library(metPath)
#> Warning in fun(libname, pkgname): mzR has been built against a different Rcpp version (1.0.5)
#> than is installed on your system (1.0.6). This might lead to errors
#> when loading mzR. If you encounter such issues, please send a report,
#> including the output of sessionInfo() to the Bioc support forum at
#> https://support.bioconductor.org/. For details see also
#> https://github.com/sneumann/mzR/wiki/mzR-Rcpp-compiler-linker-issue.
#> ✓ metID 0.9.0
#> metPath,
#> More information can be found at https://jaspershen.github.io/metPath/
#> Authors: Xiaotao Shen (shenxt@stanford.edu)
#> Maintainer: Xiaotao Shen.
#> Version 0.0.9 (20200406)Load KEGG pathway human database
data("kegg_hsa_pathway", package = "metPath")
kegg_hsa_pathway
#> ---------Pathway source&version---------
#> KEGG & 2021-03-02
#> -----------Pathway information------------
#> 342 pathways
#> 332 pathways haves genes
#> 0 pathways haves proteins
#> 280 pathways haves compounds
#> Pathway class: Metabolism; Carbohydrate metabolism;Metabolism; Lipid metabolism;Metabolism; Metabolism of cofactors and vitamins;Metabolism; Energy metabolism;Metabolism; Amino acid metabolism;Metabolism; Nucleotide metabolism;Metabolism; Biosynthesis of other secondary metabolites;Metabolism; Metabolism of other amino acids;Metabolism; Glycan biosynthesis and metabolism;Metabolism; Metabolism of terpenoids and polyketides;Genetic Information Processing; Translation;Metabolism; Xenobiotics biodegradation and metabolism;Human Diseases; Drug resistance: antineoplastic;Environmental Information Processing; Membrane transport;Genetic Information Processing; Folding, sorting and degradation;Genetic Information Processing; Transcription;Genetic Information Processing; Replication and repair;Organismal Systems; Endocrine system;Environmental Information Processing; Signal transduction;Environmental Information Processing; Signaling molecules and interaction;Organismal Systems; Immune system;Cellular Processes; Cell growth and death;Cellular Processes; Transport and catabolism;Organismal Systems; Aging;Organismal Systems; Circulatory system;Organismal Systems; Development and regeneration;Cellular Processes; Cellular community - eukaryotes;Organismal Systems; Environmental adaptation;Organismal Systems; Nervous system;Organismal Systems; Sensory system;Cellular Processes; Cell motility;Human Diseases; Endocrine and metabolic disease;Organismal Systems; Excretory system;Organismal Systems; Digestive system;Human Diseases; Neurodegenerative disease;Human Diseases; Substance dependence;Human Diseases; Infectious disease: bacterial;Human Diseases; Infectious disease: parasitic;Human Diseases; Infectious disease: viral;Human Diseases; Cancer: overview;Human Diseases; Cancer: specific types;Human Diseases; Immune disease;Human Diseases; Cardiovascular disease
get_pathway_class(kegg_hsa_pathway)
#> # A tibble: 43 x 2
#> class n
#> * <chr> <int>
#> 1 Cellular Processes; Cell growth and death 8
#> 2 Cellular Processes; Cell motility 1
#> 3 Cellular Processes; Cellular community - eukaryotes 5
#> 4 Cellular Processes; Transport and catabolism 7
#> 5 Environmental Information Processing; Membrane transport 1
#> 6 Environmental Information Processing; Signal transduction 26
#> 7 Environmental Information Processing; Signaling molecules and interact… 5
#> 8 Genetic Information Processing; Folding, sorting and degradation 7
#> 9 Genetic Information Processing; Replication and repair 7
#> 10 Genetic Information Processing; Transcription 3
#> # … with 33 more rowsPathway enrichment
We use the demo compound list from metPath.
data("query_id_kegg", package = "metPath")
query_id_kegg
#> [1] "C00164" "C00099" "C00300" "C01026" "C00122" "C00037" "C05330" "C00097"
#> [9] "C00079" "C00065" "C00188" "C00082" "C00183" "C00166" "C00163" "C00022"
#> [17] "C00213"Remove the disease pathways:
remain_idx =
kegg_hsa_pathway@pathway_class %>%
unlist() %>%
stringr::str_detect("Disease") %>%
`!`() %>%
which()
pathway_database =
filter_pathway(object = kegg_hsa_pathway, remain_idx = remain_idx)
result =
enrich_kegg(query_id = query_id_kegg,
query_type = "compound",
id_type = "KEGG",
pathway_database = pathway_database,
p_cutoff = 0.05,
p_adjust_method = "BH",
threads = 3)Check the result:
result
#> ---------Pathway database&version---------
#> KEGG & 2021-03-02
#> -----------Enrichment result------------
#> 195 pathways are enriched
#> 28 pathways p-values < 0.05
#> Glycolysis / Gluconeogenesis;Citrate cycle (TCA cycle);Pentose phosphate pathway;Pentose and glucuronate interconversions;Fructose and mannose metabolism ... (only top 5 shows)Plot to show Pathway enrichment
enrich_bar_plot(object = result)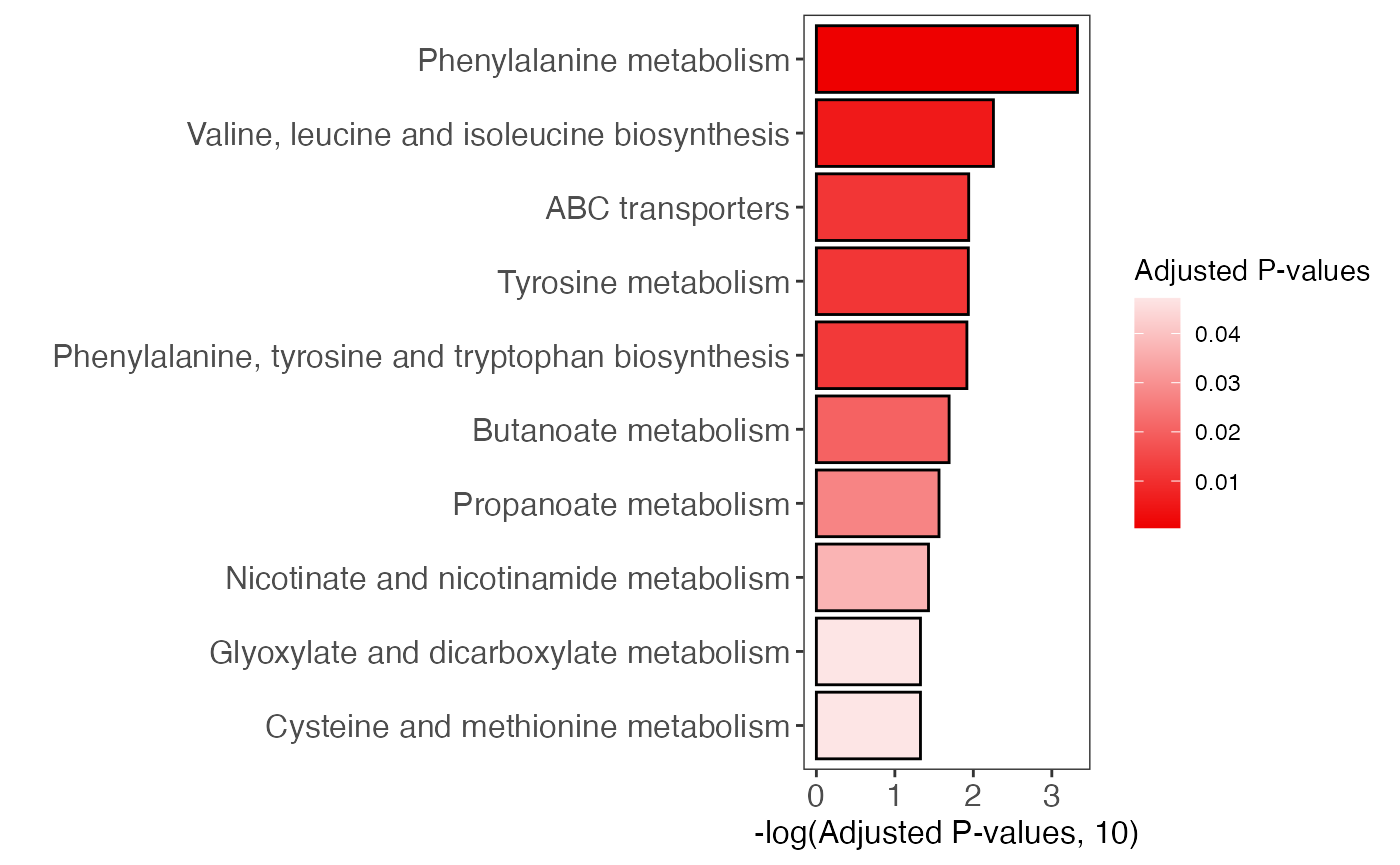
enrich_scatter_plot(object = result)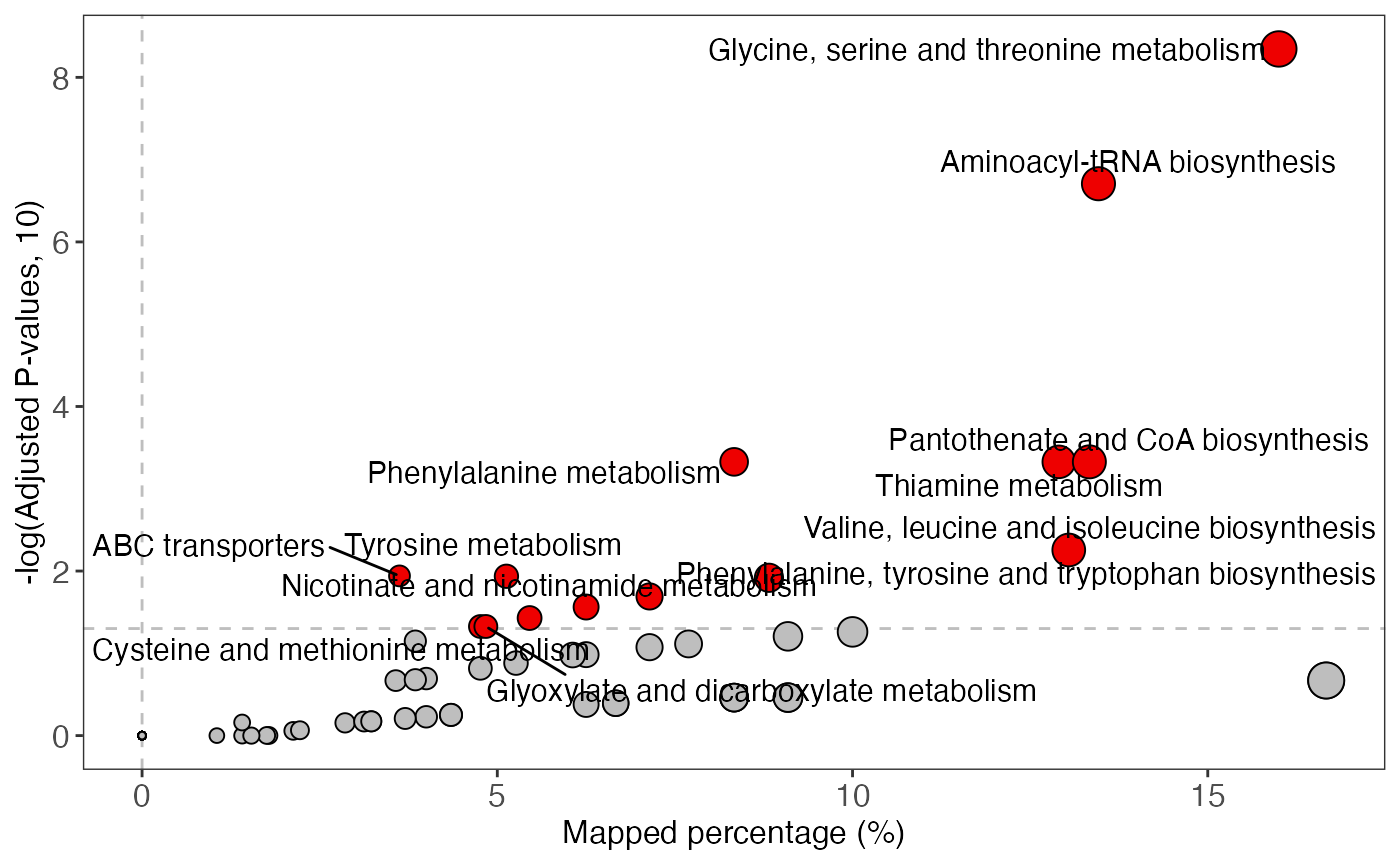
enrich_network(object = result)