HMDB pathway enrichment
Xiaotao Shen PhD (https://www.shenxt.info/)
true
Created on 2020-03-28 and updated on 2021-03-13
Source:vignettes/hmdb_pathway_enrichment.Rmd
hmdb_pathway_enrichment.RmdLoad HMDB pathway database
data("hmdb_pathway", package = "metPath")
hmdb_pathway
#> ---------Pathway source&version---------
#> SMPDB & 2021-03-02
#> -----------Pathway information------------
#> 48703 pathways
#> 0 pathways have genes
#> 48642 pathways have proteins
#> 48674 pathways have compounds
#> Pathway class: Metabolic;primary_pathway;Disease;primary_pathway;Drug Action;primary_pathway;Physiological;Physiological;primary_pathway;Drug Action;Signaling;primary_pathway;Protein;primary_pathway;Drug Metabolism;primary_pathway;Metabolic;Disease;Signaling
get_pathway_class(hmdb_pathway)
#> # A tibble: 12 x 2
#> class n
#> * <chr> <int>
#> 1 Disease 20016
#> 2 Disease;primary_pathway 232
#> 3 Drug Action 1
#> 4 Drug Action;primary_pathway 403
#> 5 Drug Metabolism;primary_pathway 64
#> 6 Metabolic 27777
#> 7 Metabolic;primary_pathway 105
#> 8 Physiological 2
#> 9 Physiological;primary_pathway 14
#> 10 Protein;primary_pathway 76
#> 11 Signaling 1
#> 12 Signaling;primary_pathway 12Pathway enrichment
We use the demo compound list from metPath.
data("query_id_hmdb", package = "metPath")
query_id_hmdb
#> [1] "HMDB0000060" "HMDB0000056" "HMDB0000064" "HMDB0000092" "HMDB0000134"
#> [6] "HMDB0000123" "HMDB0000742" "HMDB0000574" "HMDB0000159" "HMDB0000187"
#> [11] "HMDB0000167" "HMDB0000158" "HMDB0000883" "HMDB0000205" "HMDB0000237"
#> [16] "HMDB0000243" "HMDB0000271"
result =
enrich_hmdb(query_id = query_id_hmdb,
query_type = "compound",
id_type = "HMDB",
pathway_database = hmdb_pathway,
only_primary_pathway = TRUE,
p_cutoff = 0.05,
p_adjust_method = "BH",
threads = 3)Check the result:
result
#> ---------Pathway database&version---------
#> SMPDB & 2021-03-02
#> -----------Enrichment result------------
#> 874 pathways are enriched
#> 121 pathways p-values < 0.05
#> 3-Phosphoglycerate Dehydrogenase Deficiency;Dihydropyrimidine Dehydrogenase Deficiency (DHPD);Dimethylglycine Dehydrogenase Deficiency;Glycine and Serine Metabolism;Hyperglycinemia, Non-Ketotic ... (only top 5 shows)Plot to show Pathway enrichment
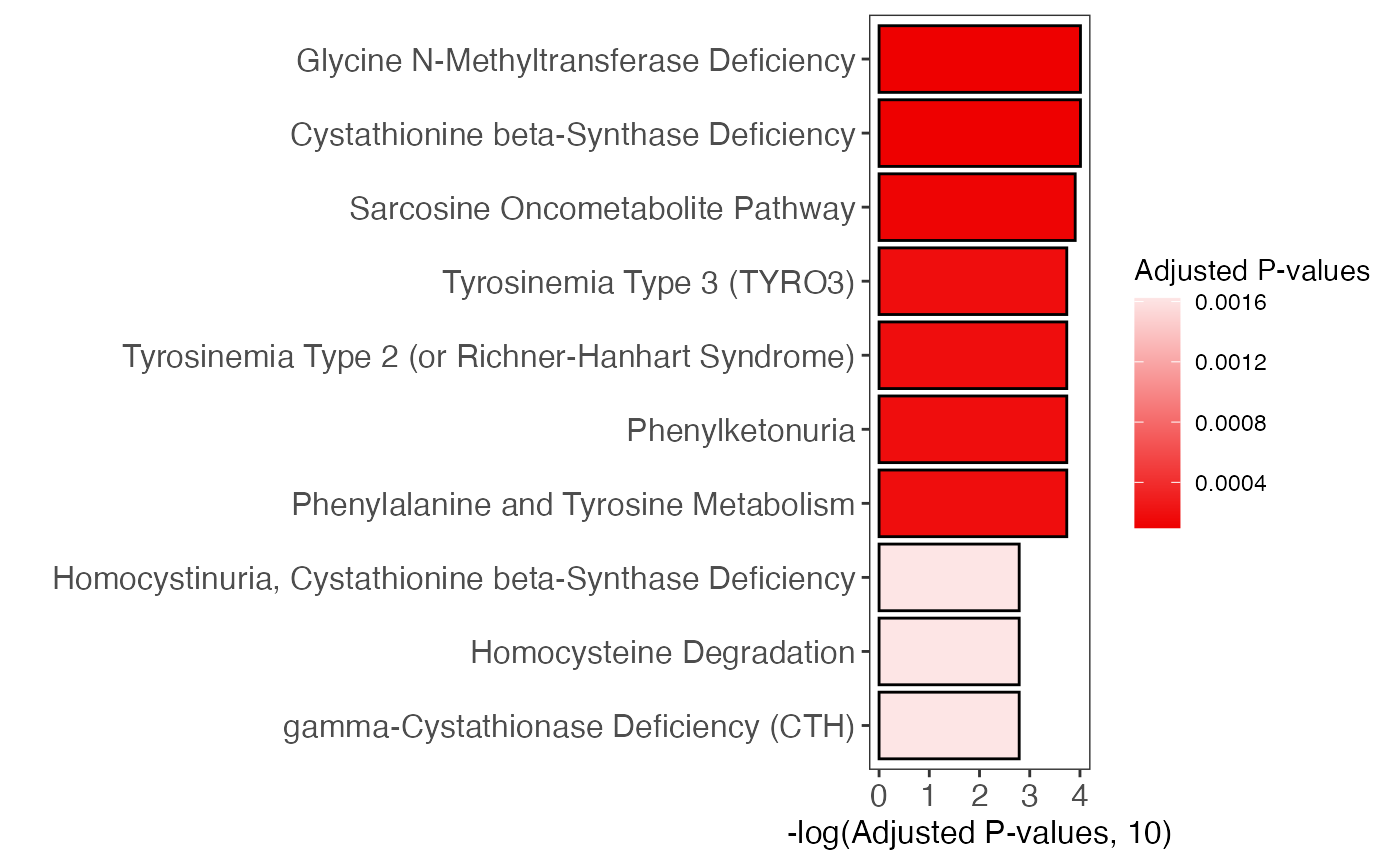
enrich_bar_plot(
object = result,
x_axis = "p_value_adjust",
cutoff = 0.05,
top = 10
)
enrich_scatter_plot(object = result)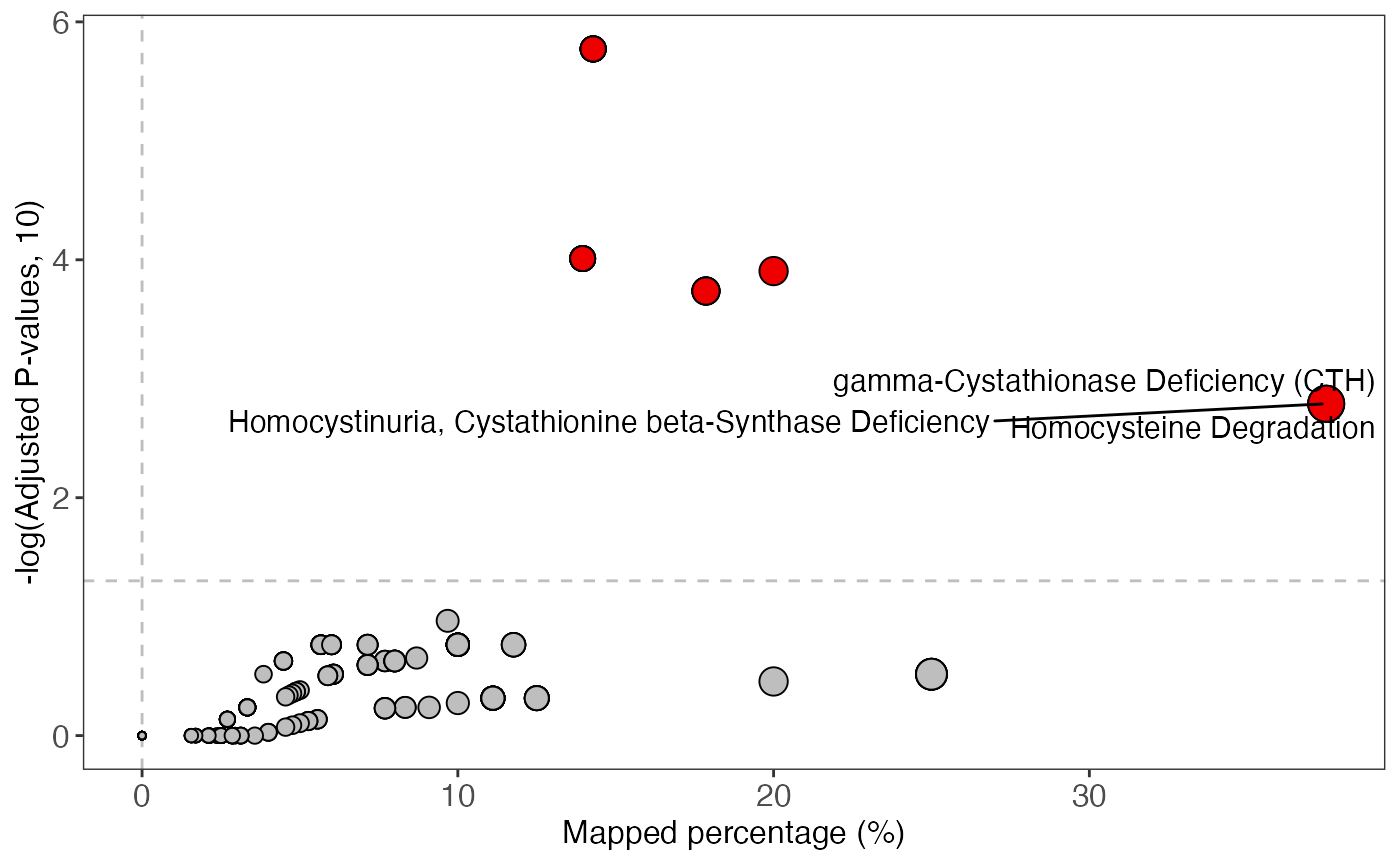
enrich_network(object = result)